Abstract
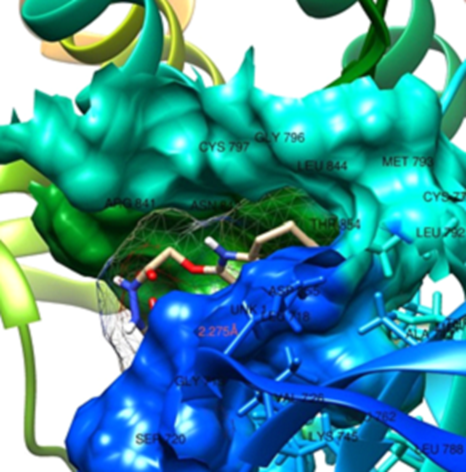
Lung carcinoma (LC) is responsible for almost one-third of all cancer fatalities worldwide. Tetrahydroquinoline is an organic molecule that is the semi-hydrogenated derivative of quinoline and could be found in several naturally occurring compounds such as flindersine, oricine etc. Some tetrahydroquinoline derivatives with pyrazole and hydrazide moieties were evaluated in silico against A549 (human lung cancer cell lines). The quantitative structural-activity relationship (QSAR) model created was statistically significant with validation metrics of R2 (0.9525), R2adj (0.9314), and CV.R (0.9719). The molecular docking analysis revealed that compound C14 demonstrated the best binding affinity towards the studied protein with binding affinity value of –10.1 kcal mol–1 (4LRM). This is in accordance with the experimental result (IC50 = 0.69). The factors observed for ADME&T correlated well with the factors observed for the referenced drug. This study indicates that compounds C1 and C9 can be further developed as anti-epidermal growth factor receptor (EGFR) compounds. Thus, our findings may open door for the design and development of library of efficient Tetrahydroquinoline-based drug-like compounds as potential anti-LC agents.
References
Adepoju, A. J.; Latona, D. F.; Olafare, O. G.; Oyebamiji, A. K.; Abdul-Hammed, M.; Semire, B. Molecular docking and pharmacokinetics studies of Curcuma longa (Curcumin) potency against Ebola virus. Ovidius University Annals of Chemistry 2022, 33 (1), 23–35. https://doi.org/10.2478/auoc-2022-0004
Banerjee, P.; Eckert, A. O.; Schrey, A. K.; Preissner, R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46 (W1), W257–W263. https://doi.org/10.1093/nar/gky318
Becke, A. D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98 (7), 5648. https://doi.org/10.1063/1.464913
Beheshti, J.; Bilal, D.; Mackey, T. P.; Limberg, L.; Bartlett, J. C.; Gwizdka, J.; Jocobson, T.; Ishimura, Y. Information literacy: Bridging the gap between theory and practice. Proceedings of the Association for Information Science and Technology, 2016, 53 (1), 1–6. https://doi.org/10.1002/pra2.2016.14505301019
Capra, J. A.; Laskowski, R. A.; Thornton, J. M.; Singh, M.; Funkhouser, T. A. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput. Biol. 2009, 5 (12), 1000585. https://doi.org/10.1371/journal.pcbi.1000585
Cassidy, A.; Myles, J. P.; van Tongeren, M.; Page, R. D.; Liloglou, T.; Duffy, S. W.; Field, J. K. The LLP risk model: An individual risk prediction model for lung cancer, Br. J. Cancer 2008, 98 (2), 270–276. https://doi.org/10.1038/sj.bjc.6604158
Cheng, A.; Dixon, S. L. In silico models for the prediction of dose-dependent human hepatotoxicity. J. Comput. Aided Mol. Des. 2003, 17 (12), 811–823. https://doi.org/10.1023/B:JCAM.0000021834.50768.c6
Collins, L. G.; Haines, C.; Perkel, R.; Enck, R. E. Lung cancer: Diagnosis and management. Am. Fam. Physician. 2007, 75 (1), 56–63.
Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. https://doi.org/10.1038/srep42717
Darvas, F.; Keseru, G.; Papp, A.; Dorman, G.; Urge, L.; Krajcsi, P. In silico and ex silico ADME approaches for drug discovery. Curr. Top. Med. Chem. 2002, 2 (12), 1287–1304. https://doi.org/10.2174/1568026023392841
Devesa, S. S.; Bray, F.; Vizcaino, A. P.; Parkin, D. M. International lung cancer trends by histologic type: Male:female differences diminishing and adenocarcinoma rates rising. Int. J. Cancer. 2005, 117 (2), 294–299 https://doi.org/10.1002/ijc.21183
Dixon, S. L.; Smondyrev, A. M.; Knoll, E. H.; Rao, S. N.; Shaw, D. E.; Friesner, R. A. PHASE: A new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J. Comput. Aided Mol. Des. 2006, 20 (10–11), 647–671. https://doi.org/10.1007/s10822-006-9087-6
Faidallah, H. M.; Rostom, S. A. F. Synthesis, in vitro antitumor evaluation and DNA binding study of novel tetrahydroquinolines and some derived tricyclic and tetracyclic ring systems. Eur. J. Med. Chem. 2013, 63, 133–143. https://doi.org/10.1016/j.ejmech.2013.02.006
Fathy, U.; Azzam, M. A.; Mahdy, F.; El-Maghraby, S.; Allam, R. M. Synthesis and in vitro anticancer activity of some novel tetrahydroquinoline derivatives bearing pyrazol and hydrazide moiety. J. Heterocycl. Chem. 2020, 57 (5), 2108–2120. https://doi.org/10.1002/jhet.3930
Grisoni, F.; Ballabio, D.; Todeschini, R.; Consonni, V. Molecular descriptors for structure–activity applications: A hands-on approach. In Computational toxicology. Nicolotti, O. Ed.; Humana Press, 2018; Vol. 1800, 3–53. https://doi.org/10.1007/978-1-4939-7899-1_1
Hayat, F.; Salahuddin, A.; Umar, S.; Azam, A. Synthesis, characterization, antiamoebic activity and cytotoxicity of novel series of pyrazoline derivatives bearing quinoline tail. Eur. J. Med. Chem. 2010, 45 (10), 4669–4675. https://doi.org/10.1016/j.ejmech.2010.07.028
Hollenberg, P. F. Characteristics and common properties of inhibitors, inducers, and activators of CYP enzymes. Drug Metab. Rev. 2002, 34 (1–2), 17–35. https://doi.org/10.1081/DMR-120001387
Ibrahim, M. T.; Uzairu, A.; Uba, S.; Shallangwa, G. A. Quantitative structure-activity relationship, molecular docking, drug-likeness, and pharmacokinetic studies of some non-small cell lung cancer therapeutic agents. Beni-Suef Univ. J. Basic Appl. Sci. 2020, 9, 49. https://doi.org/10.1186/s43088-020-00077-5
Johnson, T. O.; Adegboyega, A. E.; Iwaloye, O.; Eseola, O. A.; Plass, W.; Afolabi, B.; Rotimi, D.; Ahmed, E.; Albrakati, A.; Batiha, G. E.; Adeyemi, O. S. Computational study of the therapeutic potentials of a new series of imidazole derivatives against SARS-CoV-2. J. Pharmacol. Sci. 2021, 147 (1), 62–71. https://doi.org/10.1016/j.jphs.2021.05.004
Lapa, G. B.; Bekker, O. B.; Mirchink, E. P.; Danilenko, V. N.; Preobrazhenskaya, M. N. Regioselective acylation of congeners of 3-amino-1H-pyrazolo[3,4-b]quinolines, their activity on bacterial serine/threonine protein kinases and in vitro antibacterial (including antimycobacterial) activity. J. Enzyme Inhib. Med. Chem. 2013, 28 (5), 1088–1093. https://doi.org/10.3109/14756366.2012.716056
Lipinski, C. A.; Lombardo, F.; Dominy, B. W.; Feeney, P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46 (1–3), 3–26. https://doi.org/10.1016/S0169-409X(00)00129-0
Loving, K.; Salam, N. K.; Sherman, W. Energetic analysis of fragment docking and application to structure-based pharmacophore hypothesis generation. J. Comput. Aided Mol. Des. 2009, 23 (8), 541–554. https://doi.org/10.1007/s10822-009-9268-1
Lynch, T.; Price, A. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am. Fam. Physician. 2007, 76 (3), 391–396.
Mandewale, C. M.; Patil, U. C.; Shedge, S. V.; Dappadwad, U. R.; Yamgar, R. S. A review on quinoline hydrazone derivatives as a new class of potent antitubercular and anticancer agents. Beni-Suef Univ. J. Basic Appl. Sci. 2017, 6 (4), 354–361. https://doi.org/10.1016/j.bjbas.2017.07.005
Mekheimer, R. A.; Al-Sheikh, M. A.; Medrasi, H. Y.; Sadek, K. U. Advancements in the synthesis of fused tetracyclic quinoline derivatives. RSC Adv. 2020, 10 (34), 19867–19935. https://doi.org/10.1039/D0RA02786C
Morgensztern, D.; Ng, S. H.; Gao, F.; Govindan. R. Trends in stage distribution for patients with non-small cell lung cancer: A national cancer database survey. J. Thorac. Oncol. 2010, 5 (1), 29–33. https://doi.org/10.1097/JTO.0b013e3181c5920c
Muegge, I.; Rarey, M. Small molecule docking and scoring. In Computational chemistry review. Lipkowitz, K. B., Boyd, D. B., Eds.; Wiley Online Library, 2001; Vol. 17, 1–60. https://doi.org/10.1002/0471224413.ch1
Ntie-Kang, F. An in silico evaluation of the ADMET profile of the StreptomeDB database. SpringerPlus 2013, 2, 353. https://doi.org/10.1186/2193-1801-2-353
Oke, A. M.; Adelakun, A. O.; Akintelu, S. A.; Soetan, E. A.; Oyebamiji, A. K.; Ewemoje, T. A. Inhibition of angiotensin converting enzyme by phytochemicals in Cucurbita pepo L.: In silico Approach. Pharmacological Research - Modern Chinese Medicine. 2022, 4, 100142. https://doi.org/10.1016/j.prmcm.2022.100142
Opoku-Temeng, C.; Dayal, N.; Sooreshjani, M. A.; Sintim, H. O. 3H-pyrazolo [4, 3-f] quinoline haspin kinase inhibitors and anticancer properties. Bioorg. Chem. 2018, 78, 418–426. https://doi.org/10.1016/j.bioorg.2018.03.031
Oyebamiji, A.K.; and Semire, B. Theoretical studies of anti-corrosion properties of triphenylimidazole derivatives in corrosion inhibition of carbon steel in acidic media via DFT approach. Anal. Bioanal. Electrochem. 2018, 10 (1), 136–146.
Oyebamiji, A. K.; Fadare, O. A.; Akintelu, S. A; Semire, B. Biological studies on anthra[1,9-cd]pyrazol-6(2D)-one analogues as anti-vascular endothelial growth factor via in silico mechanisms. Chemistry Africa 2021, 4 (4), 955–963. https://doi.org/10.1007/s42250-021-00276-2
Oyeneyin, O. E.; Iwegbulam, C. G.; Ipinloju, N.; Olajide, B. F.; Oyebamiji, A. K. Prediction of the antiproliferative effects of some benzimidazolechalcone derivatives against MCF-7 breast cancer cell lines: QSAR and molecular docking studies. Org. Commun. 2022, 15 (3), 273–287. https://doi.org/10.25135/acg.oc.132.2203
Parr, R. G.; Szentpály, L.; Liu, S. Electrophilicity index. J. Am. Chem. Soc. 1999, 121 (9), 1922–1924. https://doi.org/10.1021/ja983494x
Pettersen, E. F.; Goddard, T. D.; Huang, C. C.; Couch, G. S.; Greenblatt, D. M.; Meng, E. C.; Ferrin, T. E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25 (13), 1605–1612. https://doi.org/10.1002/jcc.20084
Puzyn, T.; Leszczynski, J.; Cronin, M. Recent Advances in QSAR Studies: Methods and applications, Vol. 8; Springer Dordrecht, 2010. https://doi.org/10.1007/978-1-4020-9783-6
Sahin, S.; Benet, L. Z. The operational multiple dosing half-life: A key to defining drug accumulation in patients and to designing extended-release dosage forms. Pharm. Res. 2008, 25 (12), 2869–2877. https://doi.org/10.1007/s11095-008-9787-9
Salim, E. I.; Jazieh, A. R.; Moore, M. A. Lung cancer incidence in the Arab League countries: Risk factors and control. Asian Pacific J. Cancer Prev. 2011, 12 (1), 17–34.
Sato, T.; Watanabe, H.; Tsuganezawa, K.; Yuki, H.; Mikuni, J.; Yoshikawa, S.; Kukimoto-Niino, M.; Fujimoto, T.; Wakiyama, T. Y.; Kojim, H.; Okabe, T.; Nagano, T.; Shirouzu, M.; Yokoyama, S.; Tanaka, A.; Honma, T. Identification of novel drug-resistant EGFR mutant inhibitors by in silico screening using comprehensive assessments of protein structures. Bioorg. Med. Chem. 2012, 20 (12), 3756–3767. https://doi.org/10.1016/j.bmc.2012.04.042
Semire, B.; Mutiu, O. A.; Oyebamiji, A. K. DFT and AB INITIO methods on NMR, IR and reactivity indices of indol-3-carboxylate and indazole-3-carboxylate derivatives of cannabinoids: Comparative study. J. Phys. Theor. Chem. 2017, 13 (4), 353–377.
Siegel, R. L.; Miller, K. D.; Jemal, A. Cancer statistics, 2016. CA: Cancer. J. Clin. 2016, 66 (1), 7–30. https://doi.org/10.3322/caac.21332
Stella, G. M.; Luisetti, M.; Inghilleri, S.; Cemmi, F.; Scabini, R.; Zorzetto, M.; Pozzi, E. Targeting EGFR in non-small-cell lung cancer: Lessons, experiences, strategies. Respiratory Medicine 2012, 106 (2), 173–183. https://doi.org/10.1016/j.rmed.2011.10.015
Swierczewska, M.; Lee, K. C.; Lee, S. What is the future of PEGylated therapies? Expert Opin. Emerg. Drugs 2015, 20 (4), 531–536. https://doi.org/10.1517/14728214.2015.1113254
Testa, B.; Krämer, S. D. The Biochemistry of Drug Metabolism – An Introduction: Part 5. Metabolism and Bioactivity. Chem. Biodivers. 2009, 6 (5), 591–684. https://doi.org/10.1002/cbdv.200900022
Trott, O.; Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31 (2), 455–461. https://doi.org/10.1002/jcc.21334
Vanommeslaeghe, K.; Guvench, O.; MacKerell Jr, A. D. Molecular mechanics. Curr. Pharm. Des. 2014, 20 (20), 3281–3292. https://doi.org/10.2174/13816128113199990600
Veber, D. F.; Johnson, S. R.; Cheng, H.-Y.; Smith, B. R.; Ward, K. W.; Kopple, K. D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45 (12), 2615–2623 https://doi.org/10.1021/jm020017n
Veerasamy, R.; Rajak, H.; Jain, A.; Sivadasan, S.; Varghese, C. P.; Agrawal, R. K. Validation of QSAR models - Strategies and importance. Int. J. Drug Discov. 2011, 2 (3), 511–519.
Wang, H.; Liu, X.; Rice, S. J.; Belani, C. P. Pulmonary rehabilitation in lung cancer. PM&R 2016, 8 (10), 990–996. https://doi.org/10.1016/j.pmrj.2016.03.010
Waziri, I.; Kelani, M. T.; Oyedeji-Amusa, M. O.; Oyebamiji, A. K.; Coetzee, L.-C. C.; Adeyinka, A. S.; Muller, A. J. Synthesis and computational investigation of N,N -dimethyl-4-[(Z)-(phenylimino)methyl]aniline derivatives: Biological and quantitative structural activity relationship studies. J. Mol. Struct. 2023, 1276, 134756. https://doi.org/10.1016/j.molstruc.2022.134756
Yang, L.; Feng, J.-K.; Ren, A.-M. Theoretical studies on the electronic and optical properties of two thiophene–fluorene based π-conjugated copolymers. Polymer 2005, 46 (24), 10970–10981. https://doi.org/10.1016/j.polymer.2005.09.050
Yasuda, H.; Park, E.; Yun, C. H.; Sng, N. J.; Lucena-Araujo, A. R.; Yeo, W. L.; Huberman, M. S.; Cohen, D. W.; Nakayama, S.; Ishioka, K.; Yamaguchi, N.; Hanna, M.; Oxnard, G. R.; Lathan, C. S.; Moran, T.; Sequist, L. V.; Chaft, J. E.; Riely, G. J.; Arcila, M. E.; Soo, R. A.; Meyerson, M.; Eck, M. J.; Kobayashi, S. S.; Costa, D. B. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci. Transl. Med. 2013, 5 (216), 216ra177. https://doi.org/10.1126/scitranslmed.3007205
Zhang, Z.; Lee, J. C.; Lin, L.; Olivas, V.; Au, V.; LaFramboise, T.; Abdel-Rahman, M.; Wang, X.; Levine, A. D.; Rho, J. K.; Choi, Y. J.; Choi, C. M.; Kim, S. W.; Jang, S. J.; Park, Y. S.; Kim, W. S.; Lee, D. H.; Lee, J. S.; Miller, V. A.; Arcila, M.; Ladanyi, M.; Moonsamy, P.; Sawyers, C.; Boggon, T. J.; Ma, P. C.; Costa, C.; Taron, M.; Rosell, R.; Halmos, B.; Bivona, T. G. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet. 2012, 44 (8), 852–860. https://doi.org/10.1038/ng.2330

This work is licensed under a Creative Commons Attribution 4.0 International License.
Copyright (c) 2023 Eclética Química