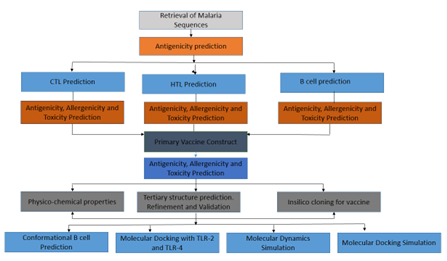
Abstract
Malaria, a life-threatening disease prevalent in tropical regions, primarily affects infants, children under five, pregnant women, travelers, and individuals with HIV/AIDS. This study utilized an immunoinformatics approach to design a peptide-based malaria vaccine targeting antigenic proteins, including Apical Membrane Antigen 1, Knob-Associated Histidine-Rich Protein, Merozoite Surface Protein 1, and Sporozoite Surface Protein 2. Antigenic protein sequences were screened for antigenicity, allergenicity, toxicity, and immune responses involving CTLs, B-cells, and HTLs. Selected epitopes were linked with appropriate linkers and an adjuvant to enhance immunogenicity, forming a vaccine construct. The construction, comprising 1473 amino acids, exhibited a molecular weight of 15.21 kDa, a theoretical pI of 8.94, an aliphatic index of 60.01, and an instability index of 31.66, indicating stability. It was hydrophilic (GRAVY: –0.385) with favorable half-lives in mammalian, yeast, and E. coli systems. Docking studies showed strong binding affinity to human TLR2 and TLR4. In silico cloning indicated a CAI value of 0.92 and a GC content of 59.31%. Further studies are needed to validate its efficacy and safety.
References
Amir, A.; Parisa, V.; Somayeh, J.; Gholamreza, A. S. A multi epitope vaccine designed against blood stage of malaria: an immunoinformatic and structural approach. Sci. Rep. 2022, 12, 11683. https://doi.org/10.1038/s41598-022-15956-3
Amlabu, E.; Mensah-Brown, H.; Nyarko, P. B.; Akuh, O. A.; Opoku, G.; Ilani, P.; Oyagbenro, R.; Asiedu, K.; Aniweh, Y.; Awandare, G. A. Functional Characterization of Plasmodium falciparum Surface-Related Antigen as a Potential Blood-Stage Vaccine Target. J Infect Dis. 2018, 218 (5), 778-790. https://doi.org/10.1093/infdis/jiy222
Biamonte, M. A.; Wanner, J.; Le Roch, K. G. Recent advances in malaria drug discovery. Bioorg. Med. Chem. 2013, 23 (10), 2829–2843. https://doi.org/10.1016/j.bmcl.2013.03.067
Chauhan,V.; Rungta, T.; Goyal, K.; Singh, M. P. Designing a multi-epitope based vaccine to combat Kaposi Sarcoma utilizing immunoinformatics approach. Sci. Rep. 2019, 9 (1), 2517. https://doi.org/10.1038/s41598-019-39299-8
Chauhan, V.; Singh, M. P. Immuno-informatics approach to design a multi-epitope vaccine to combat cytomegalovirus infection. Eur J Pharmaceut Sci. 2020, 147, 105279. https://doi.org/10.1016/j.ejps.2020.105279
Chaves, T. D. S. S.; Monteiro, W. M.; Alves, J. R.; Lacerda, M.; Lopes, M. H. Pre-travel malaria chemoprophylaxis counselling in a public travel medicine clinic in São Paulo. Brazil. Malar. J. 2017, 16, 64. https://doi.org/10.1186/s12936-017-1713-3
Crompton, P. D.; Pierce, S. K.; Miller, L. H. Advances and challenges in malaria vaccine development. J. Clin. Invest. 2010, 120 (12), 4168–4178. https://doi.org/10.1172/JCI44423
Dimitrov, I.; Bangov, I.; Flower, D. R.; Doytchinova, I. AllerTOP v. 2—a server for in silico prediction of allergens. J Mol Model. 2014, 20 (6), 2278. https://doi.org/10.1007/s00894-014-2278-5
Doytchinova, I. A.; Flower, D. R. VaxiJen. A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007, 8, 4. https://doi.org/10.1186/1471-2105-8-4
Garg, V. K.; Avashthi, H.; Tiwari, A.; Jain, P.A.; Ramkete, P. W.; Kayastha, A. M.; Singh, V. K. MFPPI–Multi FASTA ProtParam interface. Bioinformation. 2016, 12 (2), 74. https://doi.org/10.6026/97320630012074
Grote, A.; Hiller, K.; Scheer, M.; Münch, R.; Nörtemann, B.; Hempel, D. C.; Jahn, D. JCat. a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005, 33 (2), W526–W531. https://doi.org/10.1093/nar/gki376
Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. https://doi.org/10.1093/nar/gkt458
Holder, A. A. The carboxy-terminus of merozoite surface protein 1: structure, specific antibodies and immunity to malaria. Parasitology. 2009, 136 (12), 1445–1456. https://doi.org/10.1017/S0031182009990515
Ito, M.; Hayashi, K.; Minamisawa, T.; Homma, S.; Koido, S.; Shiba, K. Encryption of agonistic motifs for TLR4 into artificial antigens augmented the maturation of antigenpresenting cells. PLoS One. 2017, 12 (11), e0188934. https://doi.org/10.1371/journal.pone.0188934
Kadekoppala, M.; O'Donnell, R. A.; Grainger, M.; Crabb, B. S.; Holder, A. A. Deletion of the Plasmodium falciparum merozoite surface protein 7 gene impairs parasite invasion of erythrocytes. Eukaryot Cell. 2008, 7 (12), 2123–2132. https://doi.org/10.1128/ec.00274-08
Kalita, P.; Lyngdoh, D. L.; Padhi, A. K.; Shukla, H.; Tripathi, T. Development of multi-epitope driven subunit vaccine against Fasciolagigantica using immunoinformatics approach. Int. J. Biol. Macromol. 2019, 138, 224–233. https://doi.org/10.1016/j.ijbiomac.2019.07.024
Kristian, E. S.; Scott, E. L.; Lirong, S.; Melanie, J. S.; Anke, H.; Christine, S. H.; Ashley, M. V.; Timothy, A.S.; Robert, L. M.; Stefan, H. I. K. Interrogating the Plasmodium Sporozoite Surface: Identification of Surface-Exposed Proteins and Demonstration of Glycosylation on CSP and TRAP by Mass Spectrometry-Based Proteomics. PLOS Pathog. 2016, 12 (4), e1005606. https://doi.org/10.1371/journal.ppat.1005606
Laskowski, R. A.; MacArthur, M. W.; Thornton, J. M. https://onlinelibrary.wiley.com/ (PROCHECK: Validation of protein-structure coordinates). Chapter 25.2, 2006. https://doi.org/10.1107/97809553602060000724
Laurens, M. B. RTS, S/AS01 vaccine (Mosquirix™): an overview. Hum Vaccin Immunother. 2020, 16 (3), 480–489. https://doi.org/10.1080/21645515.2019.1669415
Lee, H. W.; Kim, M. J.; Park, M. Y.; Han, K. H.; Kim, J. The conserved proline residue in the LOB domain of LBD18 is critical for DNA-binding and biological function. Mol Plant. 2013, 6 (5), 1722–1725. https://doi.org/10.1093/mp/sst037
Li, M.; Li, M.; Lin, H.; Wang, J.; Jin, Y.; Han, F. Characterization of the novel T4-like Salmonella enterica bacteriophage STP4-a and its endolysin. Arch Virol. 2016, 161 (2), 377–384. https://doi.org/10.1007/s00705-015-2647-0
Lindner, S. E.; Miller, J. L.; Kappe, S. H. Malaria parasite pre-erythrocytic infection: preparation meets opportunity. Cell. Microbiol. 2012, 14 (3), 316–324. https://doi.org/10.1111/j.1462-5822.2011.01734.x
López-Blanco, J. R.; Aliaga, J. I.; Quintana-Ortí, E. S.; Chacón, P. iMODS. Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014, 42 (W1), W271–W276. https://doi.org/10.1093/nar/gku339
Magnan, C. N.; Zeller, M.; Kayala, M. A.; Adam, V.; Randall, A.; Felgner, P. L.; Baldi Pierre. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics. 2010, 26 (23), 2936–2943. https://doi.org/10.1093/bioinformatics/btq551
Maier, A. G.; Cooke, B. M.; Cowman, A. F.; Tilley, L. Malaria parasite proteins that remodel the host erythrocyte. Nature Reviews. Microbiology. 2009, 7 (5), 341–354. https://doi.org/10.1038/nrmicro2110
Oladipo EK, Adeniyi MO, Ogunlowo MT, Irewolede BA, Adekanola VO, Oluseyi GS, Omilola JA, Udoh AF, Olufemi SE, Adediran DA, Olonade A, Idowu UA, Kolawole OM, Oloke JK, Onyeaka H. Bioinformatics Designing and Molecular Modelling of a Universal mRNA Vaccine for SARS-CoV-2 Infection. Vaccines (Basel). 2022, 10 (12), 2107. https://doi.org/10.3390/vaccines10122107
Oladipo, E. K.; Ajayi, A. F.; Ariyo, O. E.; Onile, S. O.; Jimah, E. M.; Ezediuno, L. O.; Adebayo, O. I.; Adebayo, E. T.; Odeyemi, A. N.; Oyeleke, M. O.; Oyewole, M. P.; Oguntomi, A. S.; Akindiya, O. E.; Olamoyegun, B. O.; Aremu, V.; Arowosaye, A. O.; Aboderin, D. O.; Bello, H. B.; Senbadejo, T. Y.; Awoyelu, E. H.; Oladipo, A. A.; Oladipo, B. B.; Ajayi, L. O.; Majolagbe, O. N.; Oyawoye, O. M.; Oloke, J. K. Exploration of surface glycoprotein to design multi-epitope vaccine for the prevention of Covid-19. Inform. Med. Unlocked. 2020, 21, 100438. https://doi.org/10.1016/j.imu.2020.100438
Oladipo, E. K.; Jimah, E. M.; Irewolede, B. A.; Folakanmi, E. O.; Olubodun, O. A.; Adediran, D. A.; Akintibubo, S. A.; Odunlami, F. D.; Olufemi, S. E.; Ojo, T. O.; Akinro, O. P.; Hezekiah, O. S.; Olayinka, A. T.; Abiala, G. A.; Idowu, A. F.; Ogunniran, J. A.; Ikuomola, M. O.; Adegoke, H. M.; Idowu, U. A.; Akindiya, O. E.; Adelusi T. I. Bioinformatics analysis of structural protein to approach a vaccine candidate against Vibrio cholerae infection. Immunogenetics. 2023a, 75 (2). 99–114. https://doi.org/10.1007/s00251-022-01282-5
Oladipo, E. K.; Jimah, E. M.; Irewolede, B. A.; Folakanmi, E. O.; Olubodun, O. A.; Adediran, D. A.; Akintibubo, S. A.; Odunlami, F. D.; Olufemi, S. E.; Ojo, T. O.; Akinro, O. P.; Hezekiah, O. S.; Olayinka, A. T.; Abiala, G. A.; Idowu, A. F.; Ogunniran, J. A.; Ikuomola, M. O.; Adegoke, H. M.; Idowu, U. A.; Akindiya, O. E.; Oluwasanya, G. J.; Akanbi, G. M.; Bamigboye, F. O.; Aremu, R. O.; Awobiyi, H. O.; Kolapo, K. T.; Oluwasegun, J. A.; Olatunde, S. K.; Adelusi, T. I. Immunoinformatics design of multi-epitope peptide for the diagnosis of Schistosoma haematobium infection. J Biomol Struct Dyn. 2023b, 41 (14), 6676–6683. https://doi.org/10.1080/07391102.2022.2111358
Oladipo, E. K.; Ojo, T. O.; Olufemi, S. E.; Irewolede, B. A.; Adediran, D. A.; Abiala, A. G.; Hezekiah, O. S.; Idowu, A. F.; Oladeji, Y. G.; Ikuomola, M. O.; Olayinka, A. T.; Akanbi, G. O.; Idowu, U. A.; Olubodun, O. A.; Odunlami, F. D.; Ogunniran, J. A.; Akinro, O. P.; Adegoke, H. M.; Folakanmi, E. O.; Usman, T. A, Oladokun, E. F.; Oluwasanya, G. J.; Awobiyi, H. O.; Oluwasegun, J. A.; Akintibubo, S. A.; Jimah, E. M. Proteome based analysis of circulating SARS-CoV-2 variants: approach to a universal vaccine candidate. Genes Genomics. 2023c, 45 (12), 1489–1508. https://doi.org/10.1007/s13258-023-01426-1
Oladipo, E. K.; Adeyemo, S. F.; Akinboade, M. W.; Akinleye, T. M.; Siyanbola, K. F.; Adeogun, P. A.; Ogunfidodo, V. M.; Adekunle, C. A.; Elutade, O. A.; Omoathebu, E. E.; Taiwo, B.O.; Akindiya, E. O.; Ochola, L.; Onyeaka, H. Utilizing Immunoinformatics for mRNA Vaccine Design against Influenza D Virus. BioMedInformatics. 2024a, 4, 1572–1588. https://doi.org/10.3390/biomedinformatics4020086
Oladipo, E. K.; Ojo, T. O.; Elegbeleye, O. E.; Bolaji, O. Q.; Oyewole, M. P.; Ogunlana, A. T.; Olalekan, E. O.; Abiodun, B.; Adediran, D. A.; Obideyi, O. A.; Olufemi, S. E.; Salamatullah, A. M.; Bourhia, M.; Younous, Y. A.; Adelusi, T. I. Exploring the nuclear proteins, viral capsid protein, and early antigen protein using immunoinformatic and molecular modeling approaches to design a vaccine candidate against Epstein Barr virus. Sci. Rep. 2024b, 14 (1), 16798. https://doi.org/10.1038/s41598-024-66828-x
Oluwagbemi, O. O.; Oladipo, E. K.; Kolawole, O. M.; Oloke, J. K.; Adelusi, T. I.; Irewolede, B. A.; Dairo, E. O.; Ayeni, A. E.; Kolapo, K. T.; Akindiya, O. E.; Oluwasegun, J. A.; Oluwadara, B. F.; Fatumo, S. Bioinformatics, Computational Informatics, and Modeling Approaches to the Design of mRNA COVID-19 Candidates. Computation. 2022, 10 (7), 117. https://doi.org/10.3390/computation10070117
Pandey, K.; Sharma, M.; Saarav, I.; Singh, S.; Dutta, P.; Bhardwaj, A.; Sharma, S. Analysis of the DosRregulon genes to select cytotoxic T lymphocyte epitope, specific vaccine candidates, using a reverse vaccinology approach. Int. J. Mycobacteriol. 2016, 5 (1), 34–43. https://doi.org/10.1016/j.ijmyco.2015.10.005
Pandey, R. K.; Bhatt, T. K.; Prajapati, V. K. Novel immunoinformatics approaches to design multi-epitope subunit vaccine for malaria by investigating Anopheles salivary protein. Sci Rep. 2018, 8 (1), 1125. https://doi.org/10.1038/s41598-018-19456-1
Pandey, R. K.; Ojha, R.; Aathmanathan, V. S.; Krishnan, M.; Prajapati, V. K. Immunoinformatics approaches to design a novel multi-epitope subunit vaccine against HIV infection. Vaccine. 2018, 36 (17), 2262–2272. https://doi.org/10.1016/j.vaccine.2018.03.042
Ponomarenko, J.; Bui, H. H.; Li, W.; Fusseder, N.; Bourne, P. E.; Sette, A.; Peters, B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008, 9 (1), 514. https://doi.org/10.1186/1471-2105-9-514
Saha, S.; Raghava, G. P. S. Prediction of Continuous B-cell Epitopes in an Antigen Using Recurrent Neural Network. Proteins. 2006, 65 (1), 40–48. https://doi.org/10.1002/prot.21078
Schumacher, R. F.; Spinelli, E. Malaria in Children. J. Hematol. Infect. Dis. 2012, 4, e2012073. https://doi.org/10.4084/MJHID.2012.073
Sharma, N.; Naorem, L. D.; Jain, S.; Raghava, G. P. S. ToxinPred2: An improved method for predicting toxicity of proteins. Briefings Bioinform. 2022, 23, bbac174. https://doi.org/10.1093/bib/bbac174
Singh, B.; Kim Sung, L.; Matusop, A.; Radhakrishnan, A.; Shamsul, S. S.; Cox-Singh, J.; Thomas, A.; Conway, D. J. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004, 363 (9414), 1017–1024. https://doi.org/10.1016/S0140-6736(04)15836-4
Sinnis, P.; Zavala, F. The skin stage of malaria infection: biology and relevance to the malaria vaccine effort. Future Microbiol. 2008, 3, 275–278. https://doi.org/10.2217/17460913.3.3.275
Srinivasan, P.; Beatty, W. L.; Diouf, A.; Herrera, R.; Ambroggio, X.; Moch, J. K.; Tyler, J. S.; Narum, D. L.; Pierce, S. K.; Boothroyd, J. C.; Haynes, J. D.; Miller, L. H. Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion. Proc Natl Acad Sci. 2011, 108 (32), 13275–13280. https://doi.org/10.1073/pnas.1110303108
Tahmoorespur, M.; Nazifi, N.; Pirkhezranian, Z. In silico prediction of B-cell and T-cell epitopes of protective antigen of Bacillus anthracis in development of vaccines against anthrax. Iran J. Appl. Anim Sci. 2017, 7 (3), 429–436.
World Health Organization (WHO). World Malar. Rep. 2022, 1–186; http://www.who.int/malaria/publications/world-malaria-report-2022/report/en/
Yan, Y.; Tao, H.; He, J.; Huang, S.Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020, 15 (5), 1829–1852. https://doi.org/10.1038/s41596-020-0312-x
Zhang, W.; Liu, J.; Niu, Y.Q.; Wang, L.; Hu, X. A Bayesian regression approach to the prediction of HC-II binding affinity. Comput. Methods. Progr. Biomed. 2008, 92 (1), 1–7. https://doi.org/10.1016/j.cmpb.2008.05.002
Zhao, J. W.; Yan, M.; Shi, G.; Zhang, S. L.; Ming, L. In silico identification of cytotoxic T lymphocyte epitopes encoded by RD5 region of Mycobacterium tuberculosis. J. Inf. Dev. Countries. 2017, 11 (10), 806–810. https://doi.org/10.3855/jidc.7207
Zhou, X.; Zheng, W.; Li, Y.; Pearce, R.; Zhang, C.; Bell, E. W.; Zhang, G.; Zhang, Y I-TASSER-MTD: a deep-learning-based platform for multi-domain protein structure and function prediction. Nature Protocols. 2022, 17, 2326–2353.

This work is licensed under a Creative Commons Attribution 4.0 International License.
Copyright (c) 2025 Eclética Química